Deploying SleepFM-Clinical on Replicate: From Raw EDF Files to Clinical Predictions
Learn how we productionized the open-source SleepFM-Clinical model, built a robust inference pipeline, and deployed it on Replicate with Cog to run predictions on EDF sleep study files

Sleep is one of the richest yet most underutilized sources of physiological data in medicine. A single overnight polysomnography (PSG) study can capture brain activity, respiration, cardiac signals, and muscle tone, but turning those raw signals into actionable insight has traditionally required expert clinicians and complex pipelines.
In this blog, we’ll walk you through how we took the open-source SleepFM-Clinical foundation model, built a production-ready inference pipeline, and deployed it on Replicate using Cog, allowing anyone to run predictions directly on EDF sleep study files.
The complete pipeline looks like this:
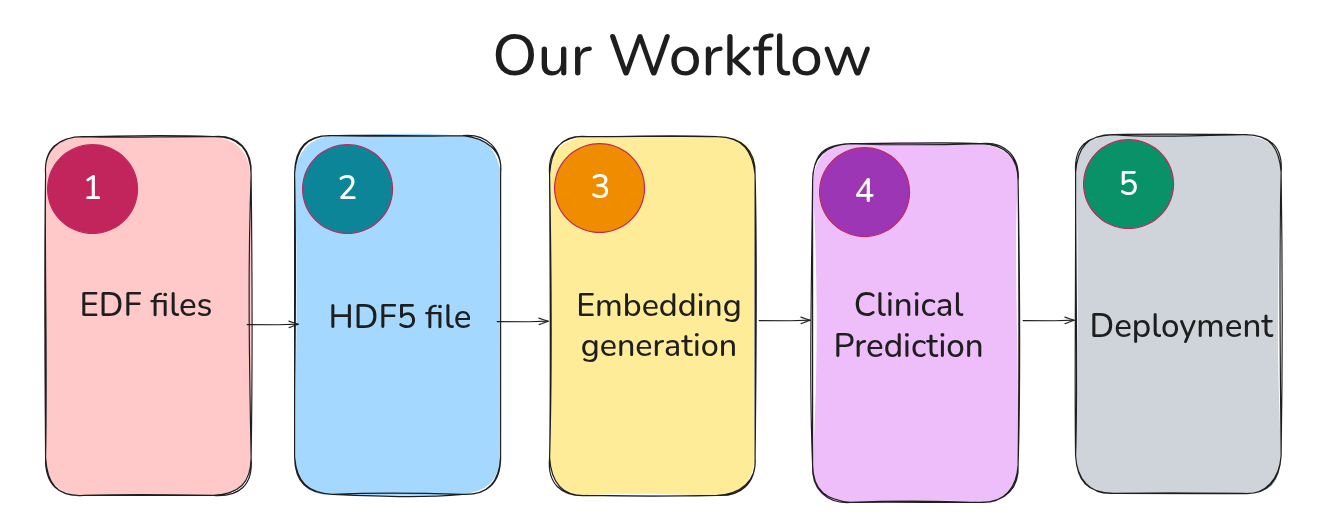
Along the way, I’ll explain:
- SleepFM architecture and how it’s different from traditional sleep models
- How Cog enables reproducible ML deployments
- How to test the model using synthetic and open-domain EDF data
Cloning the SleepFM-Clinical Repository
We begin by cloning the official repository released by the Zou Group:
Git clone git@github.com:zou-group/sleepfm-clinical.git
This repository contains:
- Pretrained SleepFM foundation models
- EDF preprocessing utilities
- Dataset abstractions
- Clinical fine-tuned models for diagnosis prediction
SleepFM is designed as a general-purpose foundation model for sleep physiology, not just sleep staging. This makes it ideal for downstream clinical tasks.
Understanding the SleepFM Model Architecture
Before touching code, it’s important to understand what SleepFM actually learns.
SleepFM is a foundation model trained on over 585,000 hours of PSG data from 65,000+ participants. It captures information present in multimodal sleep recordings, integrating EEG, ECG, EMG and respiratory signals. Its channel-agnostic architecture enables joint learning across several modalities, producing representations that generalize across environments.
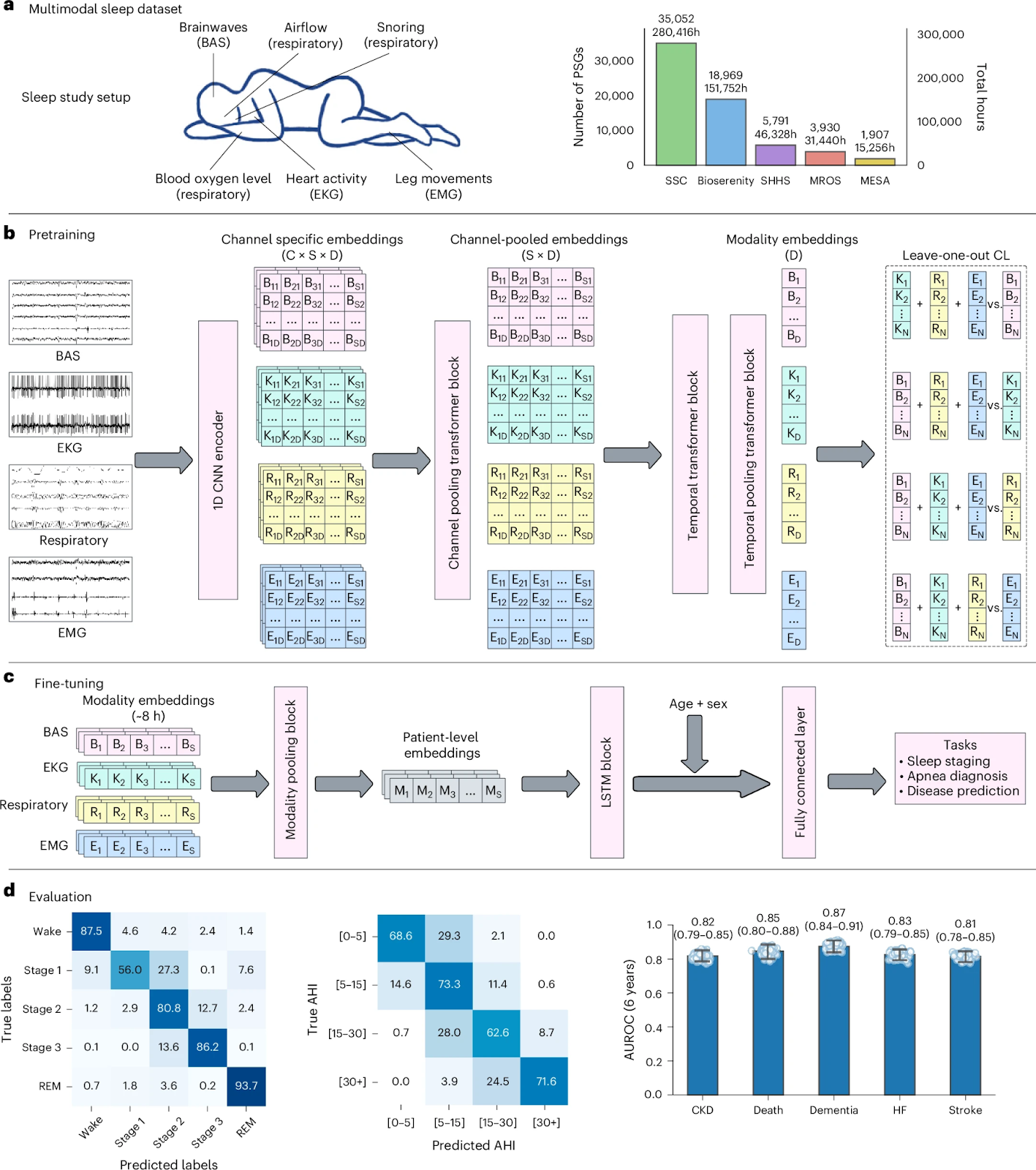
We need to remember that predicting all-cause mortality from sleep data is a particularly challenging task, as mortality risk is influenced by subtle, long-term physiological patterns rather than short-term clinical events. SleepFM substantially outperforms both a demographics-only baseline and an end-to-end PSG model trained from scratch, achieving a C-index of 0.84 (95% CI: 0.81–0.87) compared to 0.79 (0.75–0.82) for the strongest baseline.
The concordance index (C-index) measures how well a model ranks individuals by risk over time, with values closer to 1.0 indicating better discrimination. Improvements in C-index therefore reflect more accurate ordering of patients by mortality risk rather than simple classification accuracy.
Beyond overall performance, SleepFM also reveals modality-specific predictive patterns. Brain activity signals (BAS) emerge as the strongest predictors for neurological and mental health disorders, reflecting the close relationship between cortical dynamics, sleep architecture, and neuropsychiatric conditions. In contrast, respiratory signals are particularly informative for predicting senile dementia, likely due to the well-documented links between sleep-disordered breathing, intermittent hypoxia, vascular dysfunction, and neurodegeneration.
Each physiological modality captured during sleep reflects a distinct aspect of systemic health. BAS (brain activity signals) encode sleep architecture, arousal dynamics, and neural synchronization, making them particularly sensitive to neurological and psychiatric disorders. Respiratory signals reflect airflow limitation, oxygen desaturation, and breathing instability, which are strongly associated with cardiovascular risk and neurodegenerative processes. EKG signals capture autonomic nervous system activity and cardiac stress, while EMG signals provide insight into muscle tone and sleep state transitions.
By jointly modeling these modalities through a unified, channel-agnostic architecture, SleepFM integrates complementary physiological information, enabling more accurate and robust clinical predictions than any single modality alone.
What Does Channel Agnostic Mean in SleepFM ?
In SleepFM, channel agnosticism is achieved by three design choices:
(1) Per-Channel Feature Extraction
Each channel is first processed independently using 1D convolutions:
- Same convolutional encoder
- Shared weights across channels
- No channel-specific parameters
This means:
- The model does not “know” whether a signal is EEG, EKG, or RESP at this stage
- Every channel is treated as a generic physiological waveform
(2) Attention-Based Channel Pooling (Order-Invariant)
Instead of concatenating channels in a fixed order, SleepFM uses attention pooling across channels.
Key properties:
- Permutation-invariant → channel order does not matter
- Variable-size → number of channels can change
- Learned importance → model weighs informative channels more
Mathematically, this behaves like a set function, not a sequence.
So whether you pass:
[EEG, RESP, EKG]
or
[RESP, EKG]
the model can still produce a valid representation.
(3) Masking for Missing Channels
SleepFM explicitly supports channel masks:
- Missing channels are masked out
- Attention ignores masked inputs
This allows the model to:
- Train on heterogeneous cohorts
- Deploy on incomplete real-world PSGs
Modalities Used
SleepFM operates on four core physiological modalities:
- BAS (EEG-like brain activity)
- RESP (respiratory signals)
- EKG (cardiac activity)
- EMG (muscle tone)
Each modality is treated as an independent signal stream.
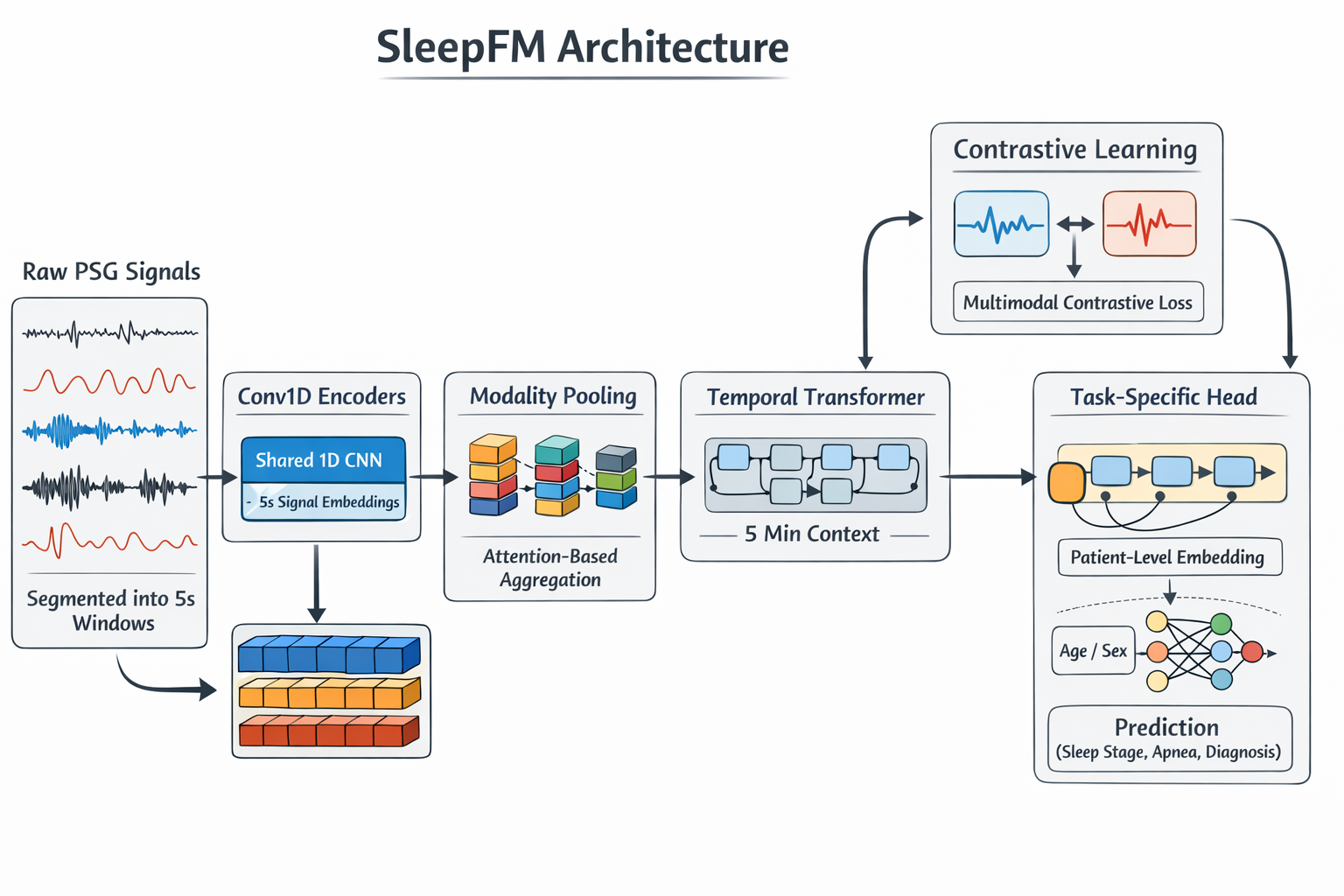
Hierarchical Embedding Generation in SleepFM
After channel-agnostic feature extraction, SleepFM transforms raw physiological signals into a hierarchy of learned embeddings that capture sleep dynamics at multiple temporal scales.
5-Second Token Embeddings: Capturing Local Physiology
All signals are first segmented into 5-second windows, which serve as the model’s fundamental input tokens. Within each window, per-channel convolutional encoders extract local temporal features, and channel-agnostic attention pooling aggregates information across available channels within each modality.
The output of this stage is a 5-second token embedding for each modality, representing short-term physiological activity such as arousals, transient breathing irregularities, or brief changes in muscle tone. These embeddings capture fine-grained, moment-to-moment dynamics in the sleep recording.
Why this matters: This temporal resolution aligns with clinically meaningful micro-events in sleep physiology while keeping the sequence length computationally manageable.
5-Minute Contextual Embeddings: Modeling Sleep Dynamics Over Time
While individual 5-second segments capture local information, many clinically relevant sleep patterns unfold over longer periods. To model these dependencies, SleepFM feeds sequences of 5-second token embeddings into a temporal transformer, which operates over a 5-minute context window.
Through self-attention, the transformer learns relationships between tokens across time, allowing the model to integrate information about sleep continuity, arousal burden, and evolving physiological states. The transformer’s output is then temporally pooled to produce a 5-minute aggregated embedding for each modality.
Why this matters: These 5-minute embeddings encode higher-level sleep structure that cannot be inferred from isolated short-term events alone.
This channel-agnostic, multimodal design is a key source of the model’s robustness, enabling it to gracefully handle missing channels and heterogeneous signal types commonly found in clinical sleep datasets.
From Embeddings to Clinical Predictions
The embeddings produced by SleepFM encode general sleep physiology and are task-agnostic. To convert these representations into clinically meaningful outputs, SleepFM applies lightweight fine-tuning models on top of the pretrained embeddings.
For downstream tasks, embeddings from all modalities (brain, respiratory, cardiac, and muscle signals) are pooled together and passed through a two-layer LSTM, which models long-range sleep dynamics across the night. For patient-level tasks such as disease or mortality prediction, all temporal information is further aggregated into a single 128-dimensional patient-level embedding, summarizing an individual’s entire sleep recording.
This compact representation is then fed into a task-specific output head (for example, disease prediction), allowing new clinical tasks to be added with minimal additional training.
Why SleepFM Is Different
Traditional sleep models:
- Are trained for a single task (e.g., sleep staging)
- Require hand-crafted features
- Break when channel layouts vary
SleepFM instead:
- Learns patch-based representations directly from raw signals
- Uses a Set Transformer architecture, making it robust to missing channels
- Produces time-aligned embeddings usable for multiple downstream tasks
This is what allows us to decouple signal understanding (foundation model) from clinical decision-making (task-specific head).
Let’s Code
Synthesize Sample EDF Files
To test the pipeline without proprietary medical data, we added a synthetic EDF generator.
First, get all the requirements using:
pip install -r requirements-dev.txt
EDF Synthesizer:
import numpy as np
import mne
channels = ['C3-A2', 'Airflow', 'Arm EMG', 'EKG']
sfreq = 256.0
duration_sec = 300
n_channels = len(channels)
n_samples = int(sfreq * duration_sec)
# Random data in safe range
data = np.random.uniform(-500, 500, size=(n_channels, n_samples))
# Create MNE info
info = mne.create_info(
ch_names=channels,
sfreq=sfreq,
ch_types=['eeg', 'misc', 'emg', 'ecg']
)
raw = mne.io.RawArray(data, info)
# Export safely
output_path = "synthetic_demo_psg2_5min.edf"
raw.export(output_path, fmt='edf', physical_range=(-500, 500))
print(f"Synthetic EDF saved at: {output_path}")
Run this file as many times; ensure you change the output path so that you don't lose any file. (Note: Ignore warnings)
Preprocess the Files
Raw EDF files are not fed directly into the model.
## table & diagram
Why Convert to HDF5?
EDF:
- Is designed for archival
- Is inefficient for chunked access
- Doesn’t store derived representations
HDF5:
- Supports chunked reads
- Is optimized for ML pipelines
- Allows incremental writes
The preprocess_edf file has two main functions:
- edf_psg()
- load_embeddings()
In edf_psg(), we take an edf file as input and then primarily perform these tasks:
- EDF-> HDF5
- Load model for embedding generation
- Create dataset and dataloader
- Run inference, generate granular(5-sec) and aggregated(5-min) embeddings
- Store them locally
def edf_psg(
edf_path: str,
base_save_path: str = "/tmp"
) -> str:
subject_id = os.path.splitext(os.path.basename(edf_path))[0]
hdf5_path = os.path.join(
base_save_path,
f"{subject_id}_psg.hdf5"
)
converter = EDFToHDF5Converter(
root_dir=base_save_path,
target_dir=base_save_path,
resample_rate=128
)
# run for single file conversion
converter.convert(edf_path, hdf5_path)
# declaring variables
model_path = "sleepfm/checkpoints/model_base"
channel_groups_path = "sleepfm/configs/channel_groups.json"
config_path = os.path.join(model_path, "config.json")
config = load_config(config_path)
channel_groups = load_data(channel_groups_path)
modality_types = config["modality_types"]
in_channels = config["in_channels"]
patch_size = config["patch_size"]
embed_dim = config["embed_dim"]
num_heads = config["num_heads"]
num_layers = config["num_layers"]
pooling_head = config["pooling_head"]
dropout = 0.0
output = os.path.join(base_save_path, "emb")
output_5min_agg = os.path.join(base_save_path, "emb_5min")
# creating directories
os.makedirs(output, exist_ok=True)
os.makedirs(output_5min_agg, exist_ok=True)
# loading model
model_class = getattr(sys.modules[__name__], config ['model'])
model = model_class(in_channels, patch_size, embed_dim, num_heads, num_layers, pooling_head=pooling_head, dropout=dropout)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# parallelizing model
if device.type == "cuda" and torch.cuda.device_count() > 1:
model = torch.nn.DataParallel(model)
model.to(device)
# loading checkpoint
checkpoint = torch.load(
os.path.join(model_path, "best.pt"),
map_location=device
)
state_dict = checkpoint["state_dict"]
if next(iter(state_dict)).startswith("module.") and not isinstance(model, torch.nn.DataParallel):
state_dict = {k.replace("module.", "", 1): v for k, v in state_dict.items()}
if not next(iter(state_dict)).startswith("module.") and isinstance(model, torch.nn.DataParallel):
state_dict = {f"module.{k}": v for k, v in state_dict.items()}
model.load_state_dict(state_dict)
model.eval()
# creating dataset
dataset = SetTransformerDataset(
config,
channel_groups,
hdf5_paths=[hdf5_path], #the single file we converted
split="test"
)
# creating dataloader
dataloader = DataLoader(
dataset,
batch_size=1,
shuffle=False,
collate_fn=collate_fn
)
# running inference
with torch.no_grad():
with tqdm.tqdm(total=len(dataloader)) as pbar:
for batch in dataloader:
batch_data, mask_list, file_paths, _, chunk_starts = batch
(bas, resp, ekg, emg) = batch_data
(mask_bas, mask_resp, mask_ekg, mask_emg) = mask_list
bas = bas.to(device, dtype=torch.float)
resp = resp.to(device, dtype=torch.float)
ekg = ekg.to(device, dtype=torch.float)
emg = emg.to(device, dtype=torch.float)
mask_bas = mask_bas.to(device, dtype=torch.bool)
mask_resp = mask_resp.to(device, dtype=torch.bool)
mask_ekg = mask_ekg.to(device, dtype=torch.bool)
mask_emg = mask_emg.to(device, dtype=torch.bool)
embeddings = [
model(bas, mask_bas),
model(resp, mask_resp),
model(ekg, mask_ekg),
model(emg, mask_emg),
]
# Model gives two kinds of embeddings. Granular 5 second-level embeddings and aggregated 5 minute-level embeddings. We save both of them below.
embeddings_new = [e[0].unsqueeze(1) for e in embeddings]
# aggregated embeddings
for i in range(len(file_paths)):
file_path = file_paths[i]
chunk_start = chunk_starts[i]
subject_id = os.path.basename(file_path).split('.')[0]
output_path = os.path.join(output_5min_agg, f"{subject_id}.hdf5")
with h5py.File(output_path, 'a') as hdf5_file:
for modality_idx, modality_type in enumerate(config["modality_types"]):
if modality_type in hdf5_file:
dset = hdf5_file[modality_type]
chunk_start_correct = chunk_start // (embed_dim * 5 * 60)
chunk_end = chunk_start_correct + embeddings_new[modality_idx][i].shape[0]
if dset.shape[0] < chunk_end:
dset.resize((chunk_end,) + embeddings_new[modality_idx][i].shape[1:])
dset[chunk_start_correct:chunk_end] = embeddings_new[modality_idx][i].cpu().numpy()
else:
hdf5_file.create_dataset(modality_type, data=embeddings_new[modality_idx][i].cpu().numpy(), chunks=(embed_dim,) + embeddings_new[modality_idx][i].shape[1:], maxshape=(None,) + embeddings_new[modality_idx][i].shape[1:])
embeddings_new = [e[1] for e in embeddings]
for i in range(len(file_paths)):
file_path = file_paths[i]
chunk_start = chunk_starts[i]
subject_id = os.path.basename(file_path).split('.')[0]
output_path = os.path.join(output, f"{subject_id}.hdf5")
# granular embeddings
with h5py.File(output_path, 'a') as hdf5_file:
for modality_idx, modality_type in enumerate(config["modality_types"]):
if modality_type in hdf5_file:
dset = hdf5_file[modality_type]
chunk_start_correct = chunk_start // (embed_dim * 5)
chunk_end = chunk_start_correct + embeddings_new[modality_idx][i].shape[0]
if dset.shape[0] < chunk_end:
dset.resize((chunk_end,) + embeddings_new[modality_idx][i].shape[1:])
dset[chunk_start_correct:chunk_end] = embeddings_new[modality_idx][i].cpu().numpy()
else:
hdf5_file.create_dataset(modality_type, data=embeddings_new[modality_idx][i].cpu().numpy(), chunks=(embed_dim,) + embeddings_new[modality_idx][i].shape[1:], maxshape=(None,) + embeddings_new[modality_idx][i].shape[1:])
pbar.update()
return os.path.join(output, f"{subject_id}.hdf5")
The other function load_embeddings() takes an HDF5 file to load generated embeddings for model prediction.
def load_embeddings(hdf5_path, max_seq_length):
"""
function to load generated embeddings from hdf5 file to be used by prediction model
"""
modalities = ["BAS", "RESP", "EKG", "EMG"] # modalities for which embeddings are generated
emb_list = []
with h5py.File(hdf5_path, "r") as f:
print(f.keys())
for m in modalities:
try:
emb = f[m][:] # (T, E) # time x embedding dimension
emb_list.append(emb)
except:
print(f"Modal {m} not found in {hdf5_path}")
# Stack → (C, T, E) # modalities x time x embedding dimension
x = np.stack(emb_list, axis=0)
# Truncate or pad sequence length
T = x.shape[1]
S = min(T, max_seq_length)
x = x[:, :S, :] # (C, S, E)
mask = np.zeros((x.shape[0], S)) # no padding
return x, mask
We are done with the most rigorous task. Now let’s get started with Cog and Replicate AI.
Wrapping Up Everything with Cog
To make the model deployable, we wrap everything inside a Cog Predictor.
Install Cog using:
sudo curl -o /usr/local/bin/cog -L https://github.com/replicate/cog/releases/latest/download/cog_`uname -s`_`uname -m`
sudo chmod +x /usr/local/bin/cog
Initialize:
cd path/to/your/model
cog init
Here we have two important files:
- cog.yaml
- predict.py
In cog.yaml:
# Configuration for Cog ⚙️
# Reference: https://cog.run/yaml
build:
# set to true if your model requires a GPU
gpu: true
# python version in the form '3.11' or '3.11.4'
python_version: "3.10"
# path to a Python requirements.txt file
python_requirements: requirements.txt
# commands run after the environment is setup
run:
- export PYTHONPATH=/src/sleepfm
# predict.py defines how predictions are run on your model
predict: "predict.py:Predictor"
The predict.py file is where we do our disease prediction. For this we have two functions: setup() and predict(), which:
- Loads our diagnosis model
- Accepts EDF FIles
- Does preprocessing and loads embeddings
- Returns predictions
All complexity stays inside the container.
- Get all the imports
from cog import BasePredictor, Input, Path
import shutil
import os
import torch
import pandas as pd
import numpy as np
from typing import Any
from sleepfm.models.models import DiagnosisFinetuneFullLSTMCOXPHWithDemo
from sleepfm.preprocessing.preprocess_edf import edf_to_psg
from sleepfm.preprocessing.preprocess_edf import load_embeddings
2. Setup function is responsible for loading the diagnosis model.
class Predictor(BasePredictor):
def setup(self) -> Any:
"""Load model + weights """
self.device = "cuda" if torch.cuda.is_available() else "cpu"
# ---- MUST match training config ----
self.embed_dim = 128
self.num_heads = 8
self.num_layers = 2
self.num_classes = 1065
self.max_seq_length = 6480
# model
self.model = DiagnosisFinetuneFullLSTMCOXPHWithDemo(
embed_dim=self.embed_dim,
num_heads=self.num_heads,
num_layers=self.num_layers,
num_classes=self.num_classes,
max_seq_length=self.max_seq_length
).to(self.device)
# checkpoint loading
checkpoint = torch.load("sleepfm/checkpoints/model_diagnosis/best.pth", map_location="cpu")
# If checkpoint was saved using DataParallel
if "module." in list(checkpoint.keys())[0]:
checkpoint = {
k.replace("module.", ""): v
for k, v in checkpoint.items()
}
self.model.load_state_dict(checkpoint)
self.model.eval()
- Predict function does the following:
- Takes EDF file as input
- Creates temporary storage for the file
- Executes edf preprocessing we’d defined
- Loads generated embeddings
- Synthesizes demographic features (age,sex): we have mean age and neutral sex
- Passes them through model
- Gets predictions
- Maps predictions to labels
def predict(
self,
edf_file: Path = Input(description="EDF sleep study file"),
) -> Any:
"""
Run prediction for ONE patient file
"""
edf_tmp_path = "/tmp/input.edf"
shutil.copy(str(edf_file), edf_tmp_path)
output_hdf5 = edf_to_psg(edf_path=edf_tmp_path)
B = 1 # batch size
C = 4 # channels (modalities)
S = self.max_seq_length # sequence length
E = self.embed_dim
with torch.no_grad():
x_np, mask_np = load_embeddings(
hdf5_path=output_hdf5,
max_seq_length=self.max_seq_length
)
# Add batch dimension
x = torch.tensor(x_np, device=self.device).unsqueeze(0) # (1, C, S, E)
mask = torch.tensor(mask_np, dtype=torch.bool, device=self.device).unsqueeze(0) # (1, C, S)
demo_features = torch.tensor(
[[0.0, 0.0]], # age at mean, gender neutral
dtype=torch.float32,
device=self.device
)
# pass data through model
outputs = self.model(x, mask, demo_features)
all_outputs = []
logits = outputs.cpu().detach().numpy()
all_outputs.append(logits)
all_outputs = np.concatenate(all_outputs, axis=0)
# mapping predictions to labels
labels_df = pd.read_csv('sleepfm/configs/label_mapping.csv')
labels_df["output"] = all_outputs[0]
prediction = labels_df.sort_values(by="output", ascending=False).head()
return {
"prediction": prediction.to_dict(orient="records")
}
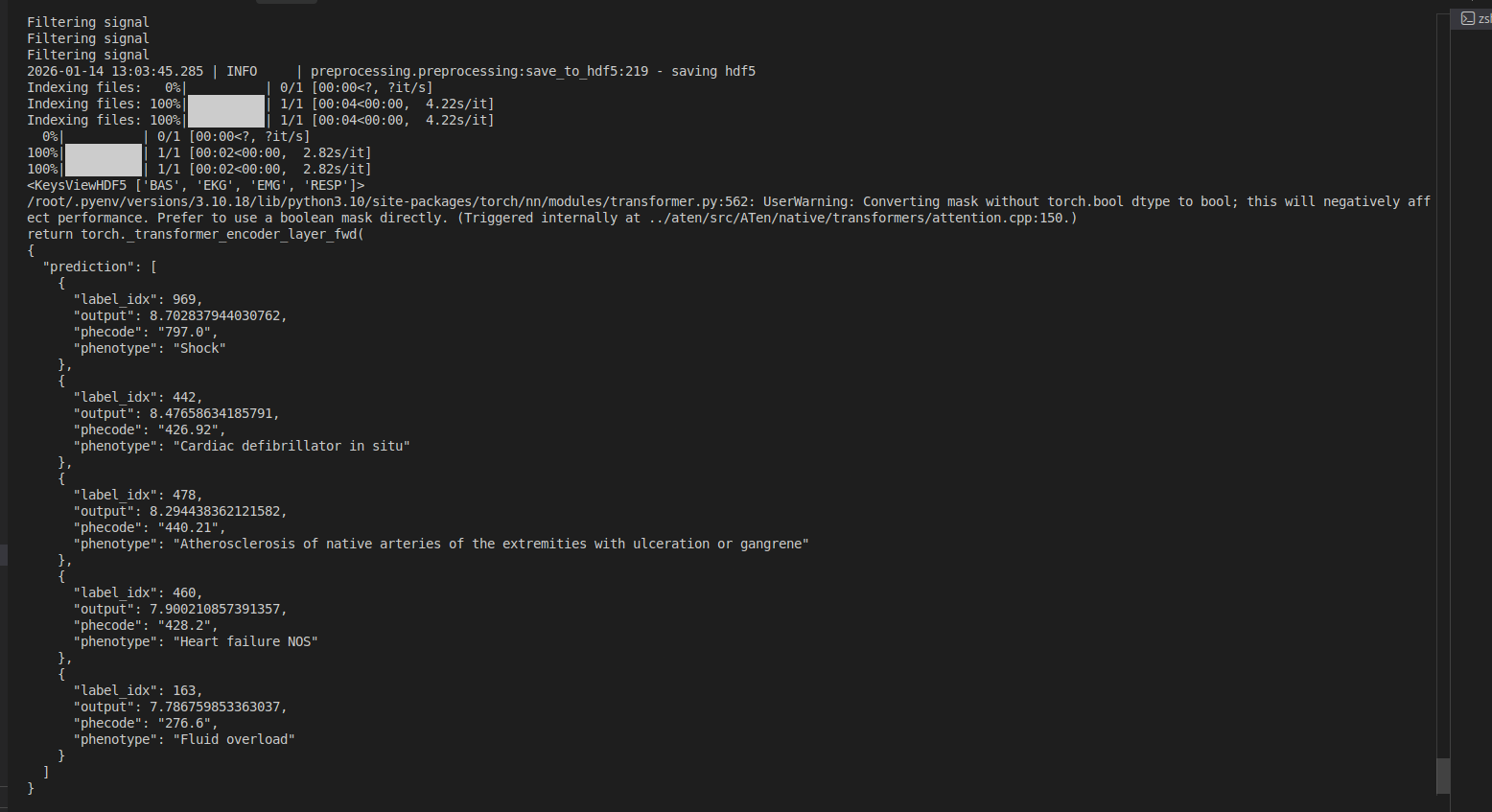
Before pushing, we can test the predict script using:
cog predict -i
edf_file=@/sleepfm-clinical/synthetic_demo_psg_5min.edf.edf # file path
Output:
Deploying to Replicate
Once the Cog setup is complete, move to Replicate AI, and add a model, then push your model. For more details refer here.
cog login
cog push r8.im/<username>/sleepfm-clinical
This creates:
- A hosted inference endpoint
- Automatic versioning
- GPU-backed execution
No custom infra required.
Testing with Replicate SDK
Using the Replicate Python SDK:
We need Replicate API, copy yours from the dashboard and export it using:
export REPLICATE_API_TOKEN=r8_******
Create a new file replicate_demo and put the code below:
import replicate
edf_path = "/sleepfm-clinical/synthetic_demo_psg2_5min.edf"
# open the file in binary mode
with open(edf_path, "rb") as f:
output = replicate.run(
"model/disease-predictor:831", # your model slug
input={"edf_file": f}
)
print("Prediction output:", output)
Output:
$ python3 replicate_demo.py
Prediction output: {'prediction': [{'label_idx': 969, 'output': 8.70824909210205, 'phecode': '797.0', 'phenotype': 'Shock'}, {'label_idx': 442, 'output': 8.481276512145996, 'phecode': '426.92', 'phenotype': 'Cardiac defibrillator in situ'}, {'label_idx': 478, 'output': 8.29833984375, 'phecode': '440.21', 'phenotype': 'Atherosclerosis of native arteries of the extremities with ulceration or gangrene'}, {'label_idx': 460, 'output': 7.910840034484863, 'phecode': '428.2', 'phenotype': 'Heart failure NOS'}, {'label_idx': 163, 'output': 7.796809673309326, 'phecode': '276.6', 'phenotype': 'Fluid overload'}]}
API Deployment (Optional)
You can go ahead and create your own deployment in your Replicate dashboard. Once you have your deployment ready, you can copy the model’s information and use the API endpoints to test it.
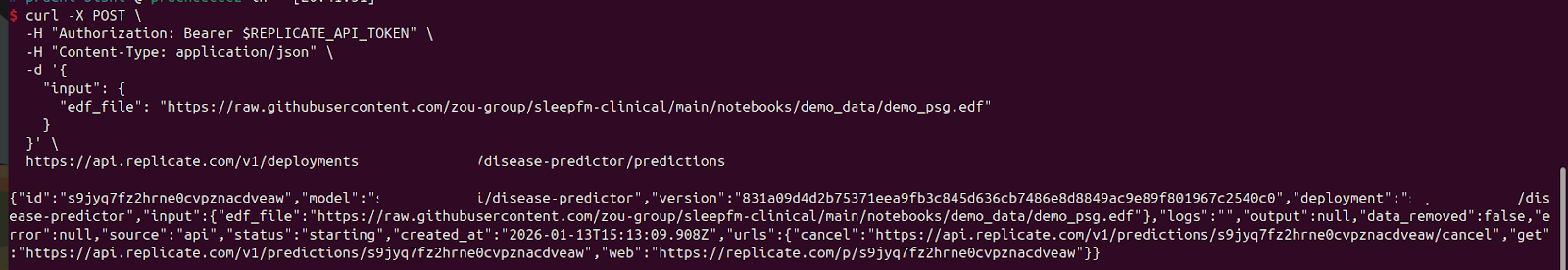
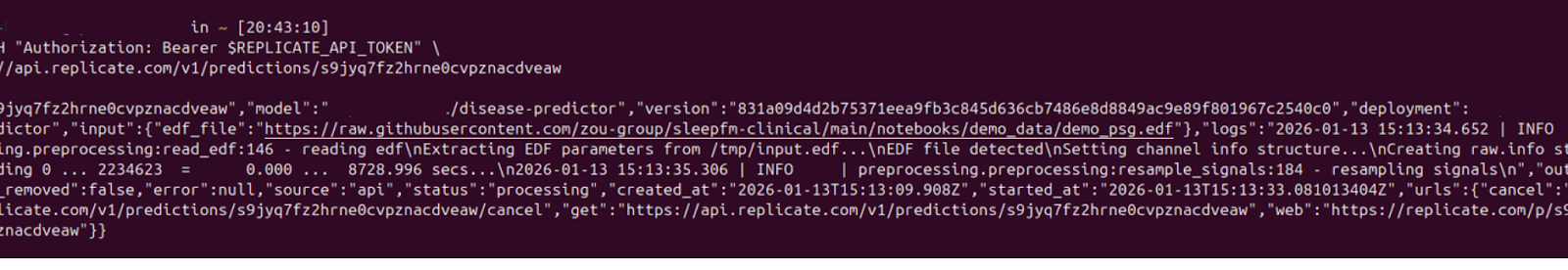
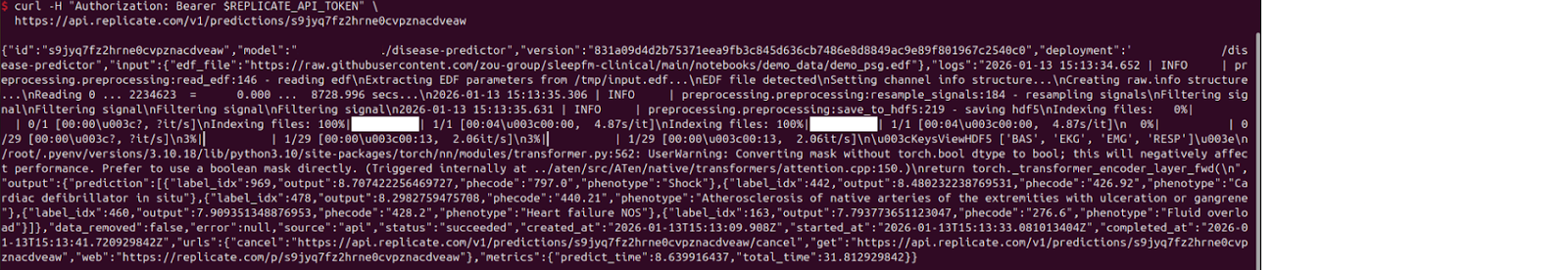
For using the API endpoints, you need your file(s) to be publicly accessible. I have used the sleepfm demo file.
curl -X POST \
-H "Authorization: Bearer $REPLICATE_API_TOKEN" \
-H "Content-Type: application/json" \
-d '{
"input": {
"edf_file": "https://raw.githubusercontent.com/zou-group/sleepfm-clinical/main/notebooks/demo_data/demo_psg.edf"
}
}' \
https://api.replicate.com/v1/deployments/model-info
Output:
Final Thoughts
SleepFM shows how large-scale, multimodal foundation models can unlock clinically meaningful insights from raw sleep data when paired with the right engineering. By building an end-to-end EDF-to-prediction pipeline and deploying it with Cog and Replicate, we make advanced sleep modeling more accessible, reproducible, and usable beyond the research setting.
At its core, SleepFM’s channel-agnostic design helps with robust learning across heterogeneous and incomplete PSG recordings, which is a critical requirement for real-world clinical data. This combination of strong representations and practical deployment gives us a scalable blueprint for translating foundation models into real clinical impact.
Ten years ago, this pipeline would have lived in a locked research lab, run once a month, and required a PhD to operate. Today, it runs on a GPU container, takes an EDF file as input, and returns clinical predictions on demand.
That shift—from bespoke research to deployable infrastructure—is the real story here.
This deployment was built with production in mind, from raw clinical data to a reproducible inference endpoint. At Superteams, this is exactly the kind of work we focus on: taking serious research models and turning them into real, deployable systems.
The complete implementation used in this blog is available on GitHub, where we extend the original SleepFM-Clinical repository with preprocessing and deployment code for EDF-based inference.
To learn more, speak to us.








